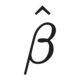Introduction
Functional genomics and bioinformatics are transforming our understanding of gene functions and interactions on a genome-wide scale. Unlike traditional genetics, which focuses on individual genes, functional genomics explores dynamic gene expression and regulation using computational and experimental approaches. Bioinformatics plays a crucial role in managing and analyzing vast genomic datasets, driving discoveries in medicine, agriculture, and biotechnology. This guide explores the methodologies, applications, computational tools, and career opportunities in this rapidly evolving field.
Functional genomics investigates how genes operate, regulate biological processes, and respond to environmental stimuli. Unlike structural genomics, which maps and sequences genes, functional genomics examines gene activity, interactions, and regulatory mechanisms. High-throughput techniques such as RNA sequencing (RNA-seq), chromatin immune precipitation sequencing (ChIP-seq), and CRISPR-based functional screens allow researchers to analyze gene expression, protein-DNA interactions, and genetic modifications. These insights drive advancements in disease research, targeted therapies, and agricultural improvements.
The development of functional genomics has been shaped by key milestones in genomics research. The Human Genome Project (1990-2003) provided the first complete human genome sequence, laying the groundwork for functional studies. The ENCODE Project (2003-Present) expanded knowledge of regulatory elements and non-coding RNAs. The advent of CRISPR-Cas9 genome editing (2012-Present) enabled precise gene modifications, revolutionizing functional studies. Advances in single-cell genomics (2010s-Present) have further refined our understanding of gene expression at the cellular level.
Technological innovations, including next-generation sequencing (NGS), artificial intelligence, and computational modeling, continue to expand the field’s potential. Functional genomics and bioinformatics offer vast opportunities in research, medicine, and biotechnology, shaping the future of genomic science.
Understanding Functional Genomics
Core Principles
Functional genomics represents a transformative approach in modern biology, integrating high-throughput technologies and computational analysis to unravel the dynamic functions of genes and their networks. By moving beyond static DNA sequences, this field investigates how genetic information flows through biological systems, driving cellular processes and phenotypic outcomes. Below we explore its core principles, interdisciplinary relationships, and scientific objectives.
Central Dogma and Functional Dynamics: the central dogma of molecular biology—defined by Francis Crick as the unidirectional flow of genetic information from DNA → RNA → protein—serves as a cornerstone for functional genomics. This principle underscores how functional genomics examines each stage of this flow:
- Transcriptional regulation: Genome-wide profiling of RNA expression (RNA-seq)
- Translational dynamics: Protein synthesis and post-translational modifications
- Feedback mechanisms: Epigenetic modifications influencing DNA accessibility
Unlike traditional genetics, which focuses on single-gene effects, functional genomics employs high-throughput methods like CRISPR screens and next-generation sequencing (NGS) to analyze entire gene networks. For example, siRNA libraries enable systematic gene knockdowns to identify functional roles across thousands of genes simultaneously.
Relationship to Other Omics Sciences
Functional genomics is closely linked to other omics disciplines, including transcriptomics, proteomics, and metabolomics. Transcriptomics examines RNA transcripts to understand gene expression, while proteomics studies protein structure and function, and metabolomics analyzes biochemical processes. By integrating data from these fields, multi-omics approaches provide a comprehensive understanding of cellular function and regulation. Systems biology plays a crucial role in this integration, modeling the complex interactions among genes, proteins, and metabolites to reveal insights into cellular mechanisms and disease pathogenesis.
Key Objectives
Essentially, the overarching goal of functional genomics is to bridge the genotype-phenotype gap, connecting genetic variations to observable traits and disease states. Researchers aim to identify regulatory mechanisms that control gene expression, investigate the effects of epigenetic modifications, and assess how environmental factors influence genomic function. By exploring these complex interactions, functional genomics enhances our understanding of disease mechanisms, aids in the development of targeted therapies, and drives innovations in personalized medicine and biotechnology.
- Decoding Gene Function – Functional genomics aims to:
- Annotate “causal roles” of genes (e.g., essentiality in cell survival)
- Characterize noncoding elements (enhancers, miRNAs) through CRISPR interference
- Bridging Genotype to Phenotype: By correlating genomic variants with molecular phenotypes (e.g., protein abundance), the field clarifies mechanisms behind traits like drug resistance or developmental disorders. For example, whole-exome sequencing identifies mutations in cardiac ion channels linked to arrhythmias.
- Advancing Precision Medicine: High-throughput screens identify therapeutic targets, such as oncogenes susceptible to inhibition, while multi-omics profiles stratify patients based on molecular subtypes.
Thus, functional genomics bridges molecular biology and systems science, leveraging genome-wide data to decode life’s complexity. By integrating multi-omics datasets and computational modeling, it illuminates disease mechanisms, evolutionary adaptations, and therapeutic opportunities—transforming biology into a predictive, network-driven discipline.
Genomics Techniques and Methods
DNA Sequencing Technologies
Next-generation sequencing (NGS) has revolutionized genomics by enabling high-throughput DNA sequencing with applications in whole-genome, exome, and targeted sequencing. These methods allow researchers to identify mutations, structural variants, and regulatory elements with unprecedented accuracy.
Applications
- Whole-genome sequencing (WGS): Comprehensive analysis of entire genomes to identify genetic variations, structural changes, and novel genes. WGS is crucial for understanding genetic disorders, evolutionary biology, and personalized medicine.
- Exome sequencing (WES): Focuses on protein-coding regions to identify mutations associated with diseases. This approach is more cost-effective than WGS and is widely used in clinical diagnostics.
Third-Generation Sequencing (Long-Read Technologies): Third-generation sequencing technologies, such as PacBio and Oxford Nanopore, provide long-read sequencing capabilities, improving genome assembly and the analysis of complex regions like repetitive sequences and structural variations.
- PacBio: Known for high accuracy in sequencing long DNA molecules, PacBio is ideal for de novo genome assembly and studying complex genomic regions.
- Oxford Nanopore Technologies (ONT): Offers real-time sequencing with ultra-long reads using portable devices like MinION. ONT is particularly useful for rapid field sequencing and studying epigenetic modifications.
These platforms provide long-read sequencing capabilities, overcoming limitations of NGS in resolving repetitive regions and structural variants.
Applications
- Structural variant detection: Long reads are essential for identifying large deletions, duplications, and inversions.
- De novo genome assembly: Enables the construction of complete genomes from scratch, especially for organisms with complex genomes.
- Epigenetic studies: Long reads can detect DNA modifications like methylation and hydroxymethylation.
- Targeted sequencing: Involves sequencing specific genes or genomic regions, often used in clinical settings to identify mutations in known disease-causing genes.
Transcriptomic Methods
RNA sequencing (RNA-Seq) is a powerful tool for quantifying gene expression levels across different conditions. This method has revolutionized transcriptomics by allowing researchers to study gene expression across the entire transcriptome.
Alternative Splicing and RNA Modifications: Advanced techniques, such as alternative splicing analysis and RNA modification profiling, help uncover post-transcriptional regulation. Techniques include:
- Long-read RNA sequencing: Platforms like PacBio and ONT enable the detection of full-length transcripts, including alternative splicing variants.
- Chemical mapping: Techniques like m6A-seq identify RNA modifications, which play crucial roles in post-transcriptional regulation.
Single-Cell RNA-Seq and Spatial Transcriptomics
- Single-Cell RNA-Seq: This technique profiles gene expression at the single-cell level, revealing cellular heterogeneity within tissues or tumors. It helps identify rare cell populations and understand developmental processes.
- Spatial Transcriptomics: By mapping gene expression back to tissue architecture, spatial transcriptomics provides spatial context to transcriptomic data. This approach is crucial for understanding cell-cell interactions and tissue organization.
Genome Editing Tools
CRISPR-Cas9 is a revolutionary genome editing tool that enables precise gene editing by introducing targeted double-strand breaks in DNA. This system has transformed functional genomics by allowing researchers to systematically study gene functions.
Applications
- Functional genomics screens: CRISPR-Cas9 is used in high-throughput screens to identify genes essential for specific biological processes or disease phenotypes.
- Disease modeling: CRISPR-Cas9 facilitates the creation of disease models by introducing disease-causing mutations into cells or organisms.
- Therapeutic development: This technology holds promise for treating genetic diseases by correcting disease-causing mutations.
- RNA Interference (RNAi): RNAi uses small interfering RNAs (siRNAs) to silence gene expression post-transcriptionally. This method is widely used in functional studies to identify gene roles.
Chromatin and Epigenomic Methods
Chromatin Immunoprecipitation Sequencing (ChIP-Seq) maps protein-DNA interactions by combining immunoprecipitation with sequencing. This technique is crucial for studying transcription factor binding sites and histone modifications.
Applications
- Transcription factor binding: Identifies genomic regions bound by specific transcription factors, revealing regulatory networks.
- Histone modification analysis: Maps histone marks like H3K4me3 and H3K27me3, which are associated with active or repressed chromatin states.
- ATAC-Seq: Assay for Transposase-Accessible Chromatin using sequencing (ATAC-Seq) identifies open chromatin regions, which are indicative of active regulatory elements like enhancers and promoters.
DNA Methylation and Histone Modifications
Techniques
- Bisulfite sequencing: Detects methylated cytosines across the genome, providing insights into epigenetic regulation.
- ChIP-Seq for histone modifications: Examines modifications like H3K9me3 and H3K27ac, which mark repressed or active chromatin, respectively.
Proteomics Integration
Mass Spectrometry-Based Protein Analysis: Mass spectrometry (MS) is a powerful tool for identifying proteins and their post-translational modifications (PTMs) at high resolution. This technique links genomic data to proteomic function by analyzing protein expression and modifications.
Proteogenomics: Proteogenomics integrates proteomics with genomics to annotate novel protein-coding regions or validate gene predictions. This field helps bridge the gap between genomic sequences and protein functions.
Emerging Technologies
Spatial genomics methods allow for tissue-level gene expression analysis, preserving spatial context in biological samples. Multimodal approaches, which simultaneously measure different molecular modalities, such as RNA, DNA, and proteins, are expanding functional genomics research.
- Multimodal Approaches: Innovations now allow simultaneous measurement of DNA, RNA, proteins, and epigenetic marks within single cells. This multimodal approach provides a comprehensive molecular snapshot of cellular states.
- Advances in Sample Preparation and Microfluidics: Improved microfluidic devices streamline single-cell isolation and multi-omics analyses while reducing sample input requirements. These advancements enhance the efficiency and sensitivity of molecular profiling.
Applications
- Single-cell analysis: Enables the study of rare cell populations and their molecular profiles.
- Clinical diagnostics: Facilitates the development of point-of-care diagnostics by miniaturizing complex molecular assays.
These cutting-edge techniques collectively empower researchers to decode the complexities of genomes, transcriptomes, proteomes, and epigenomes with unprecedented precision—paving the way for breakthroughs in biology and medicine.
Functional Genomics Analysis Approaches
Functional genomics employs a variety of analytical methods to uncover the dynamic roles of genes and their interactions within biological systems. These approaches integrate statistical, computational, and experimental techniques to interpret large-scale genomic data effectively.
Gene Expression Analysis
Differential expression analysis identifies genes whose expression levels vary significantly across different conditions (e.g., healthy vs. diseased tissues). Tools like DESeq2 and edgeR leverage statistical models to determine whether observed differences are biologically meaningful.
Applications – Identifying biomarkers, understanding disease mechanisms, and studying cellular responses to treatments.
Raw gene expression data often contain technical biases, requiring normalization techniques like TPM (Transcripts Per Million) or RPKM (Reads Per Kilobase Million) to ensure comparability across samples. Log transformations are commonly applied to stabilize variance and make data suitable for statistical analysis.
Advanced Statistical Approaches
High-dimensional gene expression datasets present challenges such as multicollinearity and sparsity. Techniques like principal component analysis (PCA) reduce dimensionality, while machine learning algorithms (e.g., random forests or support vector machines) classify patterns in complex datasets. Bayesian models are increasingly used for robust inference in noisy environments.
Network and Pathway Analysis
Gene regulatory networks (GRNs) model interactions between genes, transcription factors, and regulatory elements. Methods like ARACNe (Algorithm for the Reconstruction of Accurate Cellular Networks) infer GRNs using mutual information measures, while machine learning approaches predict regulatory relationships from transcriptomic data.
Applications: Understanding gene regulation in development, cancer progression, and immune responses.
Pathway enrichment analysis identifies biological pathways significantly associated with a set of genes or proteins. Tools like DAVID and KEGG Pathway Mapper quantify enrichment scores to highlight pathways involved in processes like metabolism or signal transduction.
Protein-Protein Interaction
Protein-protein interaction (PPI) networks map functional relationships between proteins based on experimental data (e.g., yeast two-hybrid assays) or computational predictions. PPI networks are crucial for understanding cellular machinery and identifying drug targets in diseases like cancer or neurodegeneration.
Functional Enrichment Analysis
GO enrichment analysis categorizes genes into functional groups based on shared biological processes, molecular functions, or cellular components. Tools like GOseq account for biases in gene length during enrichment calculations, providing accurate insights into gene functions.
Gene Set Enrichment Analysis (GSEA): GSEA evaluates whether predefined gene sets show statistically significant differences between conditions. Unlike traditional methods that focus on individual genes, GSEA emphasizes coordinated changes across pathways or biological processes.
Annotation-Based Tools and Databases
Functional annotation tools like Enrichr and databases such as Reactome provide curated resources for interpreting genomic data within biological contexts. These platforms integrate diverse datasets to enhance the reliability of functional interpretations.
Comparative Genomics
Comparative genomics analyzes functional elements across species to identify conserved regions critical for biological functions. Techniques like synteny mapping align genomic sequences to reveal evolutionary conservation of regulatory elements and coding regions.
Conserved Regulatory Elements: Methods such as phylogenetic footprinting identify conserved noncoding regions that likely serve regulatory roles (e.g., enhancers). These analyses help uncover universal mechanisms underlying gene regulation across diverse organisms.
Evolutionary Insights
Comparative functional genomics provides insights into how genetic variations drive phenotypic diversity and adaptation. For example, studying conserved pathways across species elucidates mechanisms of development and disease evolution.
Multi-Omics Data Integration
Statistical Methods for Integration
Integrating multi-omics data requires statistical frameworks that correlate genomic, transcriptomic, proteomic, and metabolomic datasets. Canonical correlation analysis (CCA) links datasets by identifying shared patterns, while matrix factorization methods decompose complex data into interpretable components.
Correlating Diverse Data Types: Approaches like network-based integration combine omics layers into unified models that capture interdependencies between molecular types (e.g., DNA variations influencing RNA expression or protein abundance). This enables comprehensive understanding of biological systems.
Multi-omics integration faces challenges such as batch effects, missing data, and computational complexity due to high dimensionality. Solutions include:
- Imputation techniques: Filling gaps in incomplete datasets using machine learning methods.
- Scalable algorithms: Leveraging cloud computing for large-scale analyses.
- Visualization tools: Platforms like Cytoscape simplify interpretation by creating intuitive network diagrams.
Functional genomics analysis approaches collectively enable researchers to decode complex biological systems by integrating diverse datasets with advanced computational tools—ultimately advancing our understanding of gene functions, interactions, and their implications for health and disease.
Computational Functional Genomics
Computational functional genomics integrates bioinformatics tools, algorithms, and machine learning approaches to analyze and interpret large-scale genomic datasets. It serves as the backbone for understanding gene functions, regulatory networks, and biological systems.
Bioinformatics for Functional Genomics
Bioinformatics plays a central role in functional genomics by enabling the analysis, integration, and interpretation of vast amounts of genomic, transcriptomic, proteomic, and epigenomic data. It provides computational tools to assign functional relevance to genes and their products, predict gene regulatory interactions, and model biological pathways.
Computational Infrastructure: Functional genomics requires robust computational infrastructure for data storage, processing, and analysis. This includes:
- High-performance computing (HPC): Essential for handling large-scale sequencing data.
- Cloud-based platforms: Tools like Google Cloud or AWS facilitate scalable genomic analyses.
- Databases: Resources such as Ensembl, KEGG, and InterPro provide curated datasets for annotation and pathway mapping.
Interdisciplinary Nature
Computational functional genomics is inherently interdisciplinary, combining biology, computer science, mathematics, and statistics. This integration enables researchers to develop predictive models for gene function and disease mechanisms.
Sequence Analysis Tools
Quality Control, Alignment, and Assembly Software
- Quality control: Tools like FastQC ensure the integrity of raw sequencing data.
- Alignment: Bowtie2 and BWA align reads to reference genomes efficiently.
- Assembly: SPAdes and Velvet reconstruct genomes from short reads.
Variant Calling and Annotation: Software such as GATK identifies genetic variants (SNPs and indels), while tools like ANNOVAR annotate these variants with functional information.
Specialized Algorithms
Different sequencing technologies require tailored algorithms:
- Short-read sequencing: Optimized for Illumina platforms.
- Long-read sequencing: Algorithms like Minimap2 handle PacBio or Oxford Nanopore data effectively.
Transcriptome Analysis Software
RNA-Seq Data Processing: Software like STAR or HISAT2 aligns RNA reads to reference genomes, while tools such as DESeq2 quantify gene expression levels.
Alternative Splicing Analysis: Programs like rMATS identify splicing events across conditions, revealing regulatory complexity in gene expression.
Single-Cell RNA-Seq Analysis: Specialized packages like Seurat enable single-cell RNA-Seq analysis by clustering cells based on expression profiles. Spatial transcriptomics tools add spatial context to single-cell data.
Epigenomic Data Analysis
Processing Tools for ChIP-Seq, ATAC-Seq, and Methylation Data
- ChIP-Seq: MACS identifies peaks corresponding to protein-DNA interactions.
- ATAC-Seq: Tools like HMMRATAC map accessible chromatin regions.
- Methylation analysis: Bisulfite sequencing software detects DNA methylation patterns.
Regulatory Element Identification: Motif discovery tools such as MEME identify binding motifs within regulatory regions. These motifs help predict transcription factor binding sites.
Integration with Gene Expression Data: Epigenomic data can be integrated with transcriptomic profiles using frameworks like EpiMix to explore how chromatin modifications influence gene expression patterns.
Machine Learning Applications
Machine learning methods are widely used in genomic data analysis:
- Supervised learning: Algorithms like random forests classify gene expression patterns.
- Unsupervised learning: Clustering methods (e.g., k-means) identify hidden structures in high-dimensional datasets.
Deep Learning Methods for Functional Element Prediction
Deep neural networks predict functional elements such as enhancers or splice sites by analyzing sequence features. Tools like DeepBind specialize in motif prediction using deep learning models.
AI Applications in Gene Function Prediction: AI models integrate multi-omics datasets to predict gene functions and model biological pathways. These approaches are increasingly used in drug discovery and personalized medicine.
Visualization Tools
Genome Browsers and Visualization Platforms: Genome browsers like UCSC Genome Browser or Ensembl provide interactive platforms for exploring genomic data within a biological context.
Network Visualization Tools
Cytoscape visualizes gene regulatory networks or protein-protein interaction networks, enabling researchers to interpret complex relationships between genes or proteins.
Dimensionality reduction methods like t-SNE or UMAP create intuitive visualizations of multi-layered genomic datasets (e.g., single-cell RNA-seq combined with epigenomic profiles).
Computational functional genomics continues to evolve with advancements in bioinformatics tools and machine learning algorithms. By integrating diverse datasets across omics layers, it enables deeper insights into gene functions, regulatory mechanisms, and disease biology—paving the way for breakthroughs in precision medicine and systems biology.
Conclusion
Functional genomics plays a vital role in decoding gene function by integrating large-scale genomic, transcriptomic, and proteomic data. By leveraging technologies like CRISPR screens, RNA sequencing, and chromatin mapping, researchers uncover disease mechanisms, optimize drug discovery, and enhance agricultural traits. Bioinformatics is essential in handling massive datasets and deriving meaningful biological insights, driving advancements in precision medicine, biotechnology, and environmental science.
The field is rapidly evolving with breakthroughs in single-cell sequencing, AI-driven analysis, and multi-omics integration. Future applications include personalized therapies, gene editing, and climate-resilient crops, with a growing focus on ethical considerations and data privacy. Continued innovation will expand genomics’ impact across healthcare, agriculture, and synthetic biology.
Interdisciplinary collaboration and continuous learning are key to advancing functional genomics. Researchers and professionals should engage with key databases (GenBank, Ensembl, TCGA), analysis tools (Bioconductor, Nextflow), and training programs to stay at the forefront. As technology progresses, responsible and equitable genomic research will be critical in shaping the future of medicine and biotechnology.